Hello.
Has anyone tried "NEBNext Multiplex Oligos for Illumina" to generate sequencing libraries? I have been experiencing a very poor ligation efficiency of NEBNext (hairpin) adapter. I ligated end-repaired/3'A added 1 ug of sonicated gDNA to NEBNext adapters or TruSeq adapters and amplified them by 18 cycles of PCR. The result was frustrating. The NEB one was barely amplified, whether "USER" was added or not, compared to TruSeq samples. Is there something I should've consider for the hairpin adapter ligation? If any of you has experienced this, help me please!!!
<NEBNext Oligo kit I used>
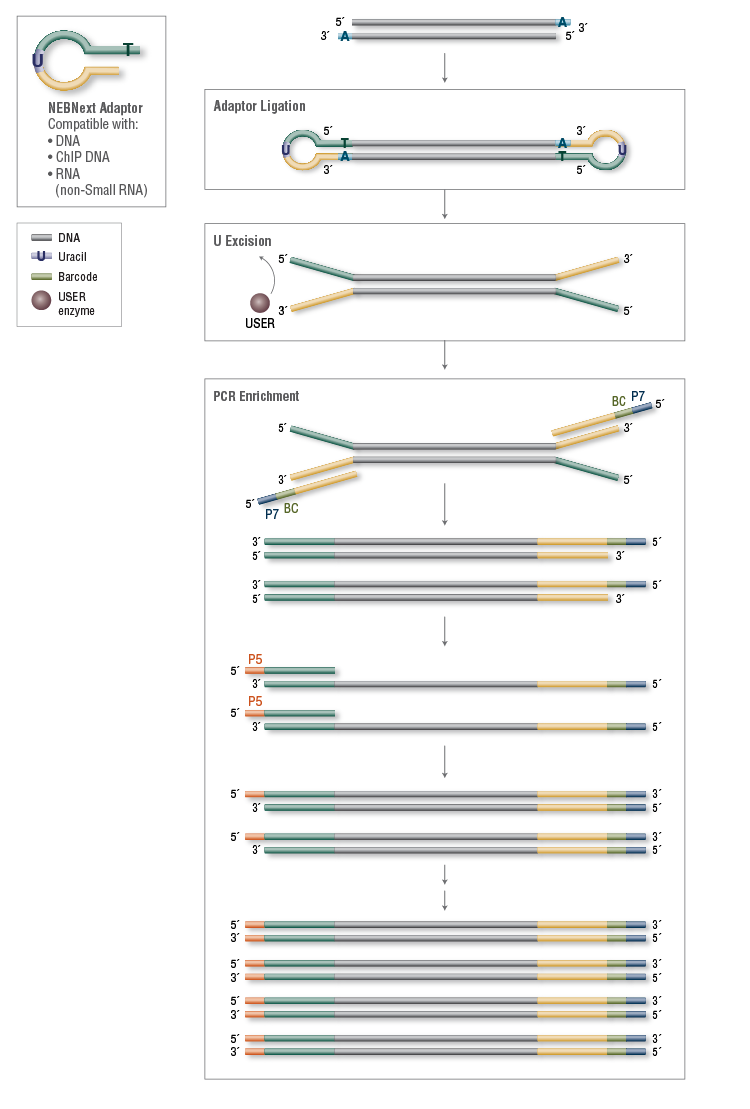
Has anyone tried "NEBNext Multiplex Oligos for Illumina" to generate sequencing libraries? I have been experiencing a very poor ligation efficiency of NEBNext (hairpin) adapter. I ligated end-repaired/3'A added 1 ug of sonicated gDNA to NEBNext adapters or TruSeq adapters and amplified them by 18 cycles of PCR. The result was frustrating. The NEB one was barely amplified, whether "USER" was added or not, compared to TruSeq samples. Is there something I should've consider for the hairpin adapter ligation? If any of you has experienced this, help me please!!!
<NEBNext Oligo kit I used>

Comment