Hello,
I am creating a database for a 2.7Gb merged sequences that I have, but I get the following error:
command used: /usr/local/bin/makeblastdb -in Merged_Lsat_1_v4.fa -dbtype nucl -parse_seqids
Building a new DB, current time: 03/04/2014 19:26:23
New DB name: Merged_Lsat_1_v4.fa
New DB title: Merged_Lsat_1_v4.fa
Sequence type: Nucleotide
Keep Linkouts: T
Keep MBits: T
Maximum file size: 1000000000B
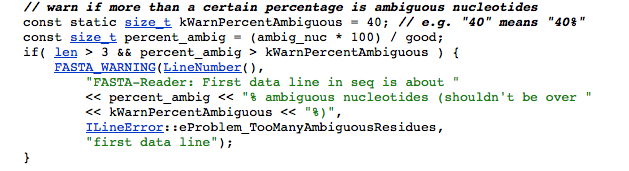
Error: (1431.1) FASTA-Reader: Warning: FASTA-Reader: First data line in seq is about 50% ambiguous nucleotides (shouldn't be over 40%)
Does anyone have a workaround or any suggestions on what I can do to fix this problem? blast doesnt quit after the error message, it just hangs in there.
Thank you,
-Milo
I am creating a database for a 2.7Gb merged sequences that I have, but I get the following error:
command used: /usr/local/bin/makeblastdb -in Merged_Lsat_1_v4.fa -dbtype nucl -parse_seqids
Building a new DB, current time: 03/04/2014 19:26:23
New DB name: Merged_Lsat_1_v4.fa
New DB title: Merged_Lsat_1_v4.fa
Sequence type: Nucleotide
Keep Linkouts: T
Keep MBits: T
Maximum file size: 1000000000B
Error: (1431.1) FASTA-Reader: Warning: FASTA-Reader: First data line in seq is about 50% ambiguous nucleotides (shouldn't be over 40%)
Does anyone have a workaround or any suggestions on what I can do to fix this problem? blast doesnt quit after the error message, it just hangs in there.
Thank you,
-Milo

Comment