Cutting DNA into small fragments is a key preparation step for DNA sequencing with NGS technology. To reduce errors and increase reliability of the sequence information, every genomic region should be sequenced several times. This means that several copies of a target DNA have to be cutted in different ways to produce overlapping fragments to ensure an good coverage of the whole region of interest. This approach is based on the general idea that genomic DNA break-points are random and sequence-independent.
But there is a problem... read more
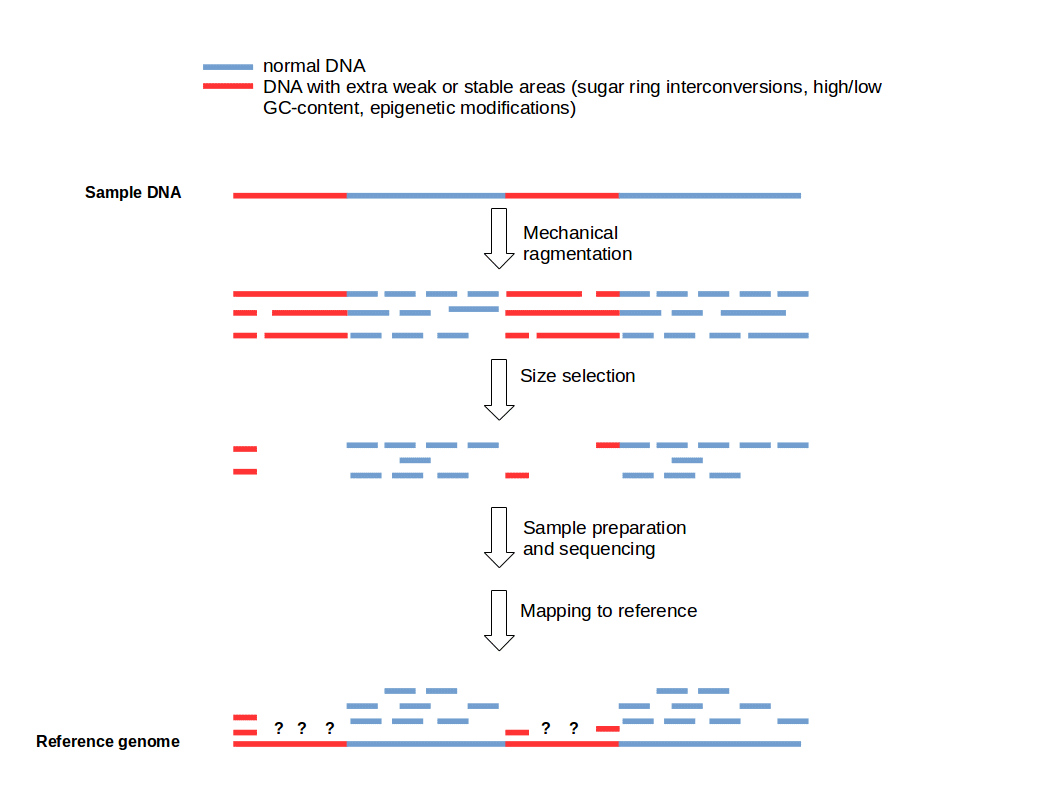
But there is a problem... read more
Comment