I want to produce a heat-map using a costumed gene list (accession IDs, e.g AT1G73590). I made a list of genes ordered depending on the processes they are involve in. I would like to conserve this order when I am plotting the expression value in csHeatmap(), but they end up ordered alphabetically.
Then I recover the FPKM from cuffdiff using getGenes
I plot the expression data using csHeatmap()
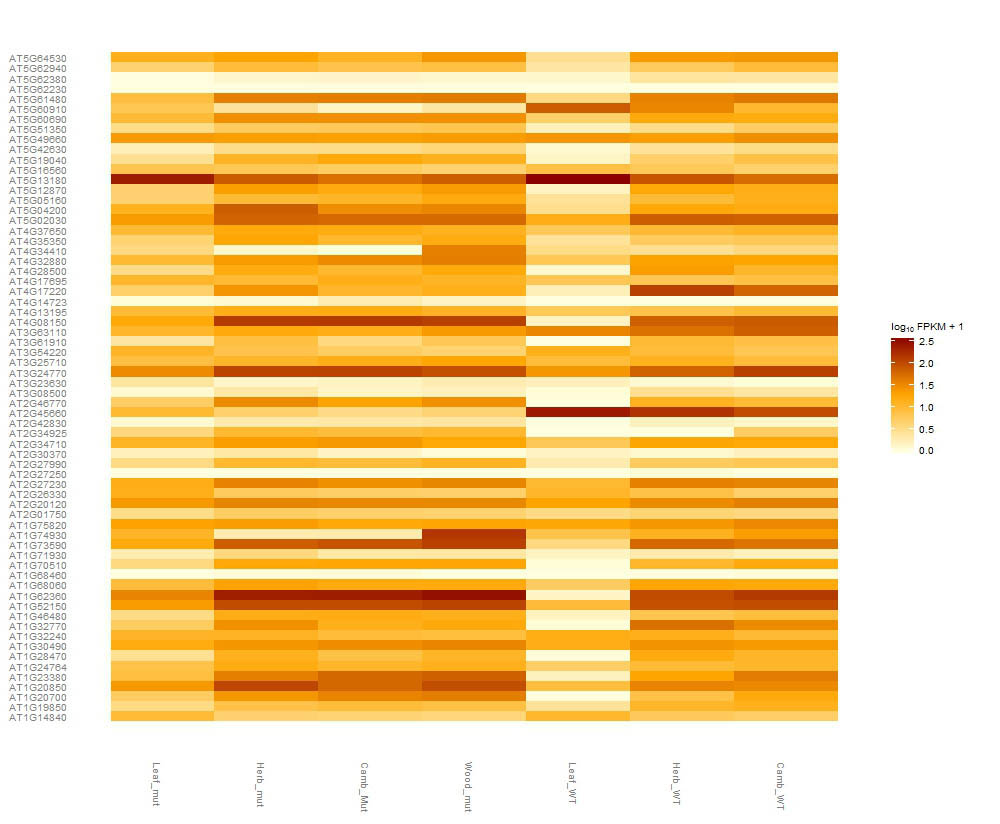
The genes are ordered alphabetically and did not keep the order my list. Any thought?
Code:
Cand_GenesIds= as.character(Gene_name_ID[,3]) head(Cand_GenesIds) [1] "AT1G19850" "AT1G73590" "AT4G32880" "AT5G16560" "AT1G32240" "AT4G17695"
Code:
Can_Genes<-getGenes(cuff, Cand_GenesIds)
Code:
can.rep<-csHeatmap(Can_Genes,cluster="none",replicates=F, fullnames=F) can.rep
The genes are ordered alphabetically and did not keep the order my list. Any thought?
Comment