Dear All,
I have a question regarding bioanalyzer for ChIPed DNA.
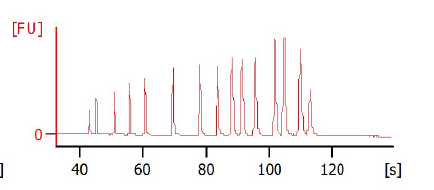
When I do bioanalyzer for the input DNA, I get peaks at the proper range in bioanalyzer.
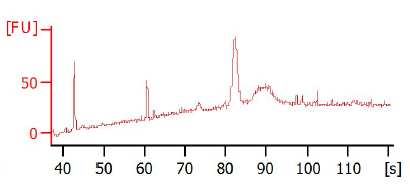
But, for the immunoprecipitated samples, I got no peaks at all or strange peaks. I even do not have the marker peaks!
I should mention that the concentration of ChIPed DNA was as low as 0.3 ng/µl (measured by Qubit HS kit) and I used 1 µl for bioanalyzer. So, I expected very low signals in bioanalyzer, but literally got nothing.
These samples were loaded in a same bioanalyzer chip used for some other library prepped samples and they all worked well, which shows that there was no problem in bioanalyzer chip or experiment.
Can anyone help me how to explain this?
I am totally confused and cannot figure out what I should do!
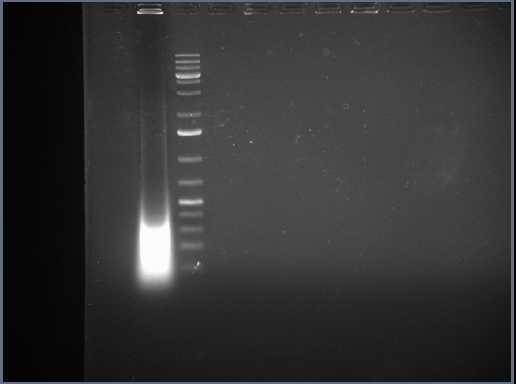
In the attached you can find a gel photo of the sonicated chromatin as well as two bioanalyzer graphs of two different ChIPed samples.
waiting for your replies...
I have a question regarding bioanalyzer for ChIPed DNA.
When I do bioanalyzer for the input DNA, I get peaks at the proper range in bioanalyzer.
But, for the immunoprecipitated samples, I got no peaks at all or strange peaks. I even do not have the marker peaks!
I should mention that the concentration of ChIPed DNA was as low as 0.3 ng/µl (measured by Qubit HS kit) and I used 1 µl for bioanalyzer. So, I expected very low signals in bioanalyzer, but literally got nothing.
These samples were loaded in a same bioanalyzer chip used for some other library prepped samples and they all worked well, which shows that there was no problem in bioanalyzer chip or experiment.
Can anyone help me how to explain this?
I am totally confused and cannot figure out what I should do!
In the attached you can find a gel photo of the sonicated chromatin as well as two bioanalyzer graphs of two different ChIPed samples.
waiting for your replies...
Comment