Introduction
MutScan is a open-source tool to detect target mutations by scanning FastQ files directly. Features:
Ultra sensitive, guarantee that all reads supporting the mutations will be detected
50X+ faster than normal pipeline (i.e. BWA + Samtools + GATK/VarScan/Mutect).
Very easy to use and need nothing else. No alignment, no reference genome, no variant call, no...
Contains built-in most actionable mutation points for cancer, like EGFR p.L858R, BRAF p.V600E...
Beautiful and informative HTML report with informative pileup visualization.
Multi-threading support.
Supports both single-end and pair-end data.
For pair-end data, MutScan will try to merge each pair, and do quality adjustment and error correction.
Able to scan the mutations in a VCF file, which can be used to visualize called variants.
Can be used to filter false-positive mutations. i.e. MutScan can handle highly repetive sequence to avoid false INDEL calling.
Application scenarios:
you are interested in some certain mutations (like cancer drugable mutations), and want to check whether the given FastQ files contain them.
you have no enough confidence with the mutations called by your pipeline, so you want to visualize and validate them to avoid false positive calling.
you worry that your pipeline uses too strict filtering and may cause some false negative, so you want to check that in a fast way.
you want to visualize the called mutation and take a screenshot with its clear pipeup information.
you called a lot of INDEL mutations, and you worry that mainly they are false positives (especially in highly repetive region)
you want to validate and visualize every record in the VCF called by your pipeline.
...
Get MutScan
MutScan on github: https://github.com/OpenGene/MutScan
Here is a sample HTML report generated my MutScan: http://opengene.org/MutScan/report.html
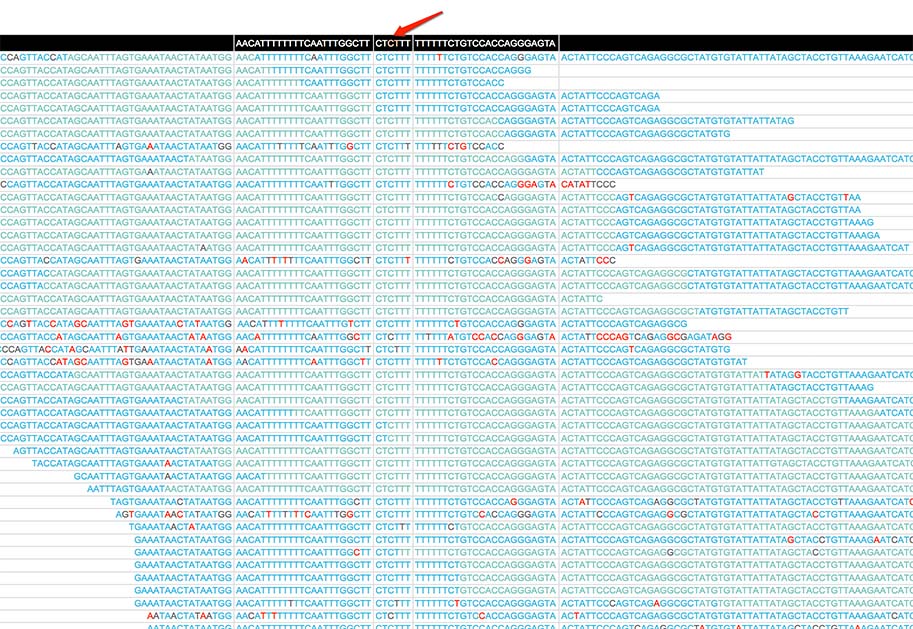
And here is a screenshot for the pileup of a mutation generated by MutScan:
MutScan is a open-source tool to detect target mutations by scanning FastQ files directly. Features:
Ultra sensitive, guarantee that all reads supporting the mutations will be detected
50X+ faster than normal pipeline (i.e. BWA + Samtools + GATK/VarScan/Mutect).
Very easy to use and need nothing else. No alignment, no reference genome, no variant call, no...
Contains built-in most actionable mutation points for cancer, like EGFR p.L858R, BRAF p.V600E...
Beautiful and informative HTML report with informative pileup visualization.
Multi-threading support.
Supports both single-end and pair-end data.
For pair-end data, MutScan will try to merge each pair, and do quality adjustment and error correction.
Able to scan the mutations in a VCF file, which can be used to visualize called variants.
Can be used to filter false-positive mutations. i.e. MutScan can handle highly repetive sequence to avoid false INDEL calling.
Application scenarios:
you are interested in some certain mutations (like cancer drugable mutations), and want to check whether the given FastQ files contain them.
you have no enough confidence with the mutations called by your pipeline, so you want to visualize and validate them to avoid false positive calling.
you worry that your pipeline uses too strict filtering and may cause some false negative, so you want to check that in a fast way.
you want to visualize the called mutation and take a screenshot with its clear pipeup information.
you called a lot of INDEL mutations, and you worry that mainly they are false positives (especially in highly repetive region)
you want to validate and visualize every record in the VCF called by your pipeline.
...
Get MutScan
MutScan on github: https://github.com/OpenGene/MutScan
Here is a sample HTML report generated my MutScan: http://opengene.org/MutScan/report.html
And here is a screenshot for the pileup of a mutation generated by MutScan:
