Here is what I mean:
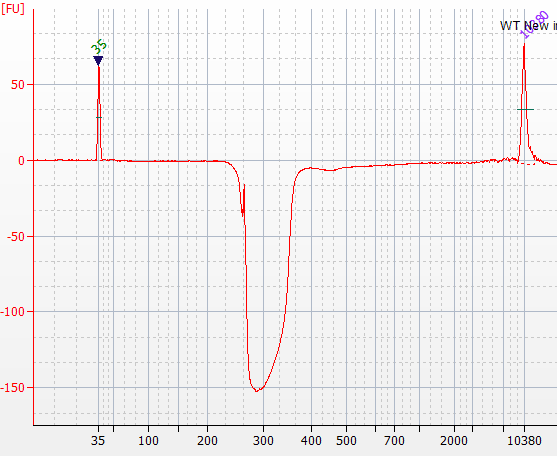
This is some input DNA obtained by reverse-crosslinking sonicated chromatin submitted by someone interested in our doing library construction for them. It was run on an Agilent Bioanalyzer High Sensitivity Chip.
Everything looks great except -- the peak produced has negative amplitude!
The Agilent Bioanalyzer Trouble shooting manual suggests this could happen as a result of the sample containing "detergents" or "dyes".
Anyone seen this and been able to figure out the cause?
(Yes, I know I answered the post of someone asking nearly the same question 4 years ago. But having seen this a few time recently in our lab, I'm finding all the responses in that thread, including my own, unhelpful.)
--
Phillip
This is some input DNA obtained by reverse-crosslinking sonicated chromatin submitted by someone interested in our doing library construction for them. It was run on an Agilent Bioanalyzer High Sensitivity Chip.
Everything looks great except -- the peak produced has negative amplitude!
The Agilent Bioanalyzer Trouble shooting manual suggests this could happen as a result of the sample containing "detergents" or "dyes".
Anyone seen this and been able to figure out the cause?
(Yes, I know I answered the post of someone asking nearly the same question 4 years ago. But having seen this a few time recently in our lab, I'm finding all the responses in that thread, including my own, unhelpful.)
--
Phillip
Comment