We are making ds-cDNA for automated mRNA-seq library preparation using the SPRIworks robot. After the poly-A selection, our protocol calls for fragmentation at 80 degrees for five minutes using ambion reagents. We get a nice bump on the bioanalyzer when running the ds-cDNA, however the peak is around 400-500 bp (see attached traces). Using the standard illumina protocol, fragmenting for 2 minutes at 94 degrees, results in a broader around 150 bp.
We plan to use the SPRIworks size selection of 200-400 bp. I am worried that after adapter ligation, much of our sample will fall outside of this range.
Any opinions? Should I just leave it alone?
We need to get some samples submitted to the robot by tomorrow, so I am planning to add 90 seconds onto the 5 minute fragmentation time. Is this a bad idea?
5 minute, 80 degree fragmentation:
[ATTACH]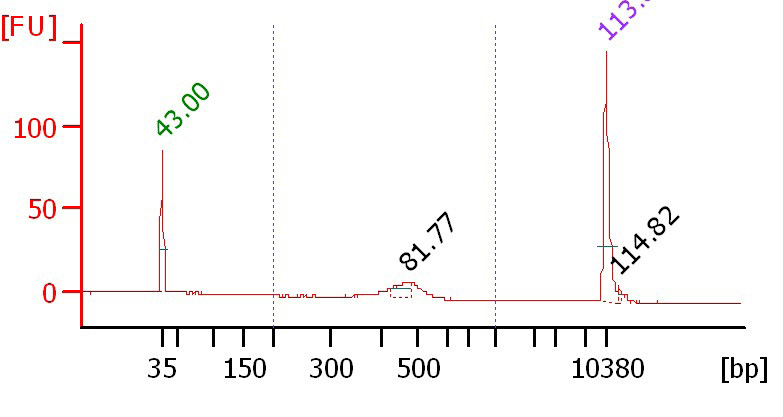 [/ATTACH]
[/ATTACH]
2 minute, 94 degree, illumina kit fragmentation:
We plan to use the SPRIworks size selection of 200-400 bp. I am worried that after adapter ligation, much of our sample will fall outside of this range.
Any opinions? Should I just leave it alone?
We need to get some samples submitted to the robot by tomorrow, so I am planning to add 90 seconds onto the 5 minute fragmentation time. Is this a bad idea?
5 minute, 80 degree fragmentation:
[ATTACH]
2 minute, 94 degree, illumina kit fragmentation: