Hi everyone,
I am glad to let you know about the official release of BioTuring Browser, the platform to resolve major challenges in single-cell RNA-seq data analysis.
With the proliferation of the single-cell RNA-seq data, we are now facing a major bottleneck in data analysis, because such enormous amounts of data require really hard code to process, and a strong computer to visualize thousands to millions of data points (i.e. thousands to millions of cells).
With BioTuring Browser, we aim to make single-cell RNA-seq data analysis extremely easy and compact for every biologist. The package can run well on a standard laptop, show up in an interactive interface that is simple to use, and provide all the state-of-the-art analyses for scRNA-seq data.
Here's to list a few features in BBrowser:
Future enhancements will include pseudotime trajectory reconstruction, the ability to search for similar single cell populations, and to combine them into the current study.
Also, BBrowser provides a rich library of scRNA-seq datasets from latest published studies, all processed and ready for visualization and reanalysis. The purpose is to remove any barriers to access valuable published data, and make science completely transparent.
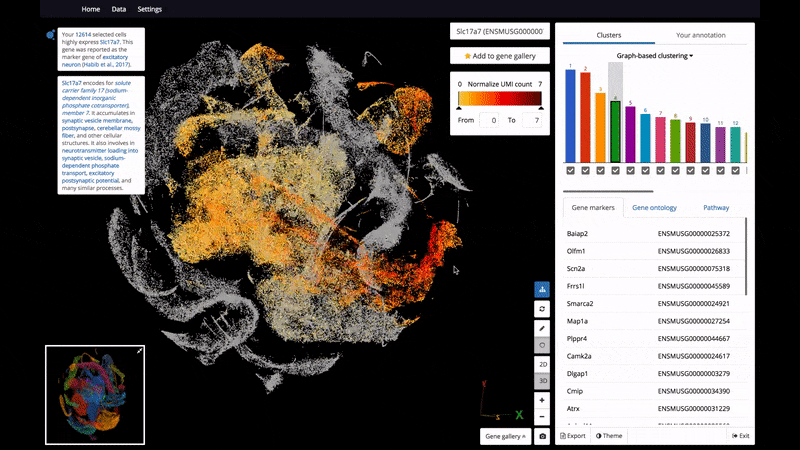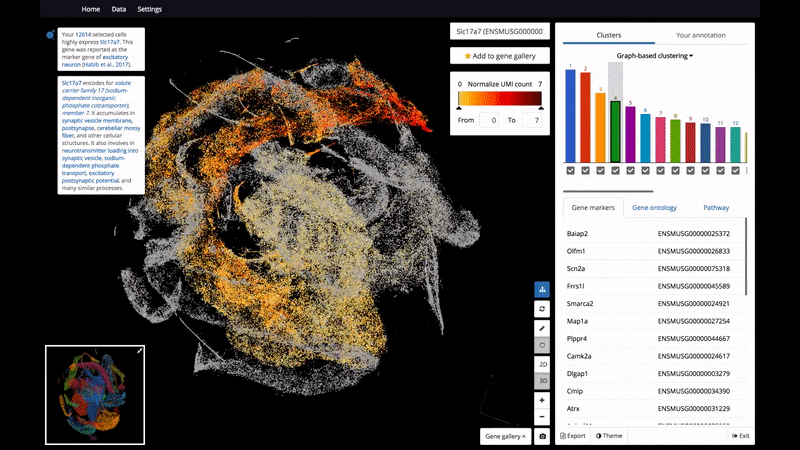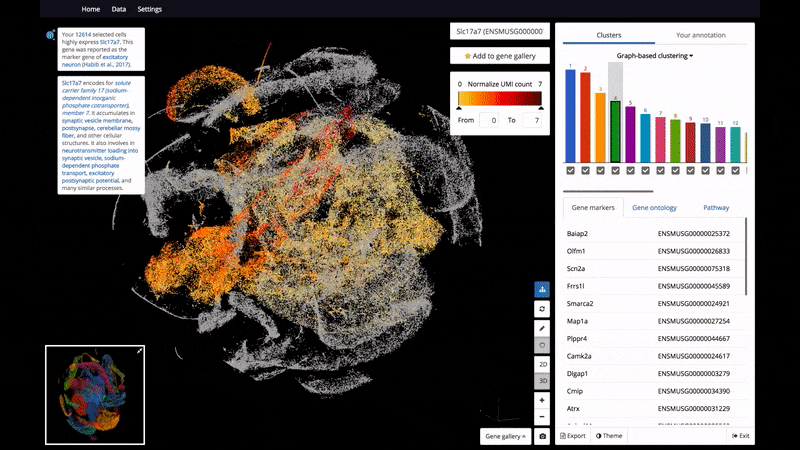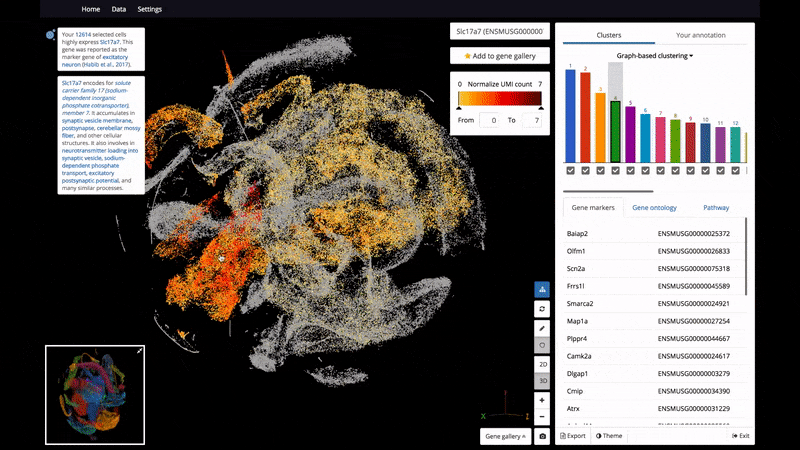
As a start, BioTuring Browser features nearly 400 publications, including almost all the recent single-cell RNA sequencing datasets and nearly 16,500 samples from bulk RNA sequencing.
BBrowser is available for Windows, Mac, and Linux at: https://bioturing.com
Feel free to raise your questions. If you are interested, please subscribe to be the first to hear about BioTuring Browser official release.
I am glad to let you know about the official release of BioTuring Browser, the platform to resolve major challenges in single-cell RNA-seq data analysis.
With the proliferation of the single-cell RNA-seq data, we are now facing a major bottleneck in data analysis, because such enormous amounts of data require really hard code to process, and a strong computer to visualize thousands to millions of data points (i.e. thousands to millions of cells).
With BioTuring Browser, we aim to make single-cell RNA-seq data analysis extremely easy and compact for every biologist. The package can run well on a standard laptop, show up in an interactive interface that is simple to use, and provide all the state-of-the-art analyses for scRNA-seq data.
Here's to list a few features in BBrowser:
- Interactive 3D/2D visualizations of up to 1 million cells on a standard laptop
- Real-time cell type prediction
- Batch effect removal (https://www.youtube.com/watch?v=0N7vouZqEoo&t=2s)
- Differential expression analysis
- Cell population comparison
- The ability to select and label cell clusters for further annotation
- Sub-clustering
- View 2 genes' expression at the same time
- View box plots/ violin plots of the expression levels across all the groups
Future enhancements will include pseudotime trajectory reconstruction, the ability to search for similar single cell populations, and to combine them into the current study.
Also, BBrowser provides a rich library of scRNA-seq datasets from latest published studies, all processed and ready for visualization and reanalysis. The purpose is to remove any barriers to access valuable published data, and make science completely transparent.

As a start, BioTuring Browser features nearly 400 publications, including almost all the recent single-cell RNA sequencing datasets and nearly 16,500 samples from bulk RNA sequencing.
BBrowser is available for Windows, Mac, and Linux at: https://bioturing.com
Feel free to raise your questions. If you are interested, please subscribe to be the first to hear about BioTuring Browser official release.
Comment