for example, there are two gene expression microarray datasets,
samples 1
gene 14 year-old 25 year-old .....
LDH 0.2 0.8 ....
samples 2
gene 12 year-old 20year-old .....
LDH 0.25 0.78
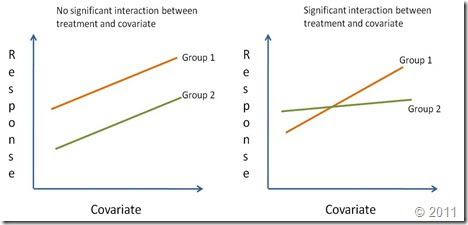
if i know the LDH gene expression level is increasing with age, I want to know the correlataion of the gene expression change in aging process between two samples, how i calculated the Pearson correlation. and how I make a ANCOVA in the above LDH expression between the two samples.
samples 1
gene 14 year-old 25 year-old .....
LDH 0.2 0.8 ....
samples 2
gene 12 year-old 20year-old .....
LDH 0.25 0.78
if i know the LDH gene expression level is increasing with age, I want to know the correlataion of the gene expression change in aging process between two samples, how i calculated the Pearson correlation. and how I make a ANCOVA in the above LDH expression between the two samples.

Comment