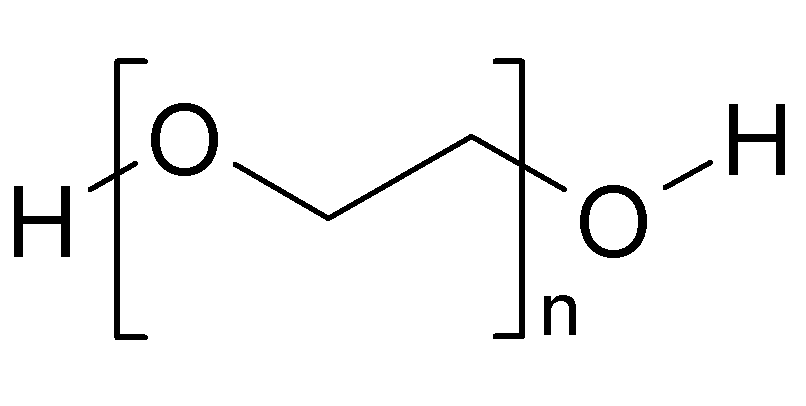
We, and our NGS service providers are frequently asked about a common clean up / size selection step during library preparation. Specifically, how does PEG precipitate DNA on negatively charged, carboxylated Ampure beads. Thought it would be useful to share our answer here:
- Genohub
- Genohub
Comment