Hello everyone,
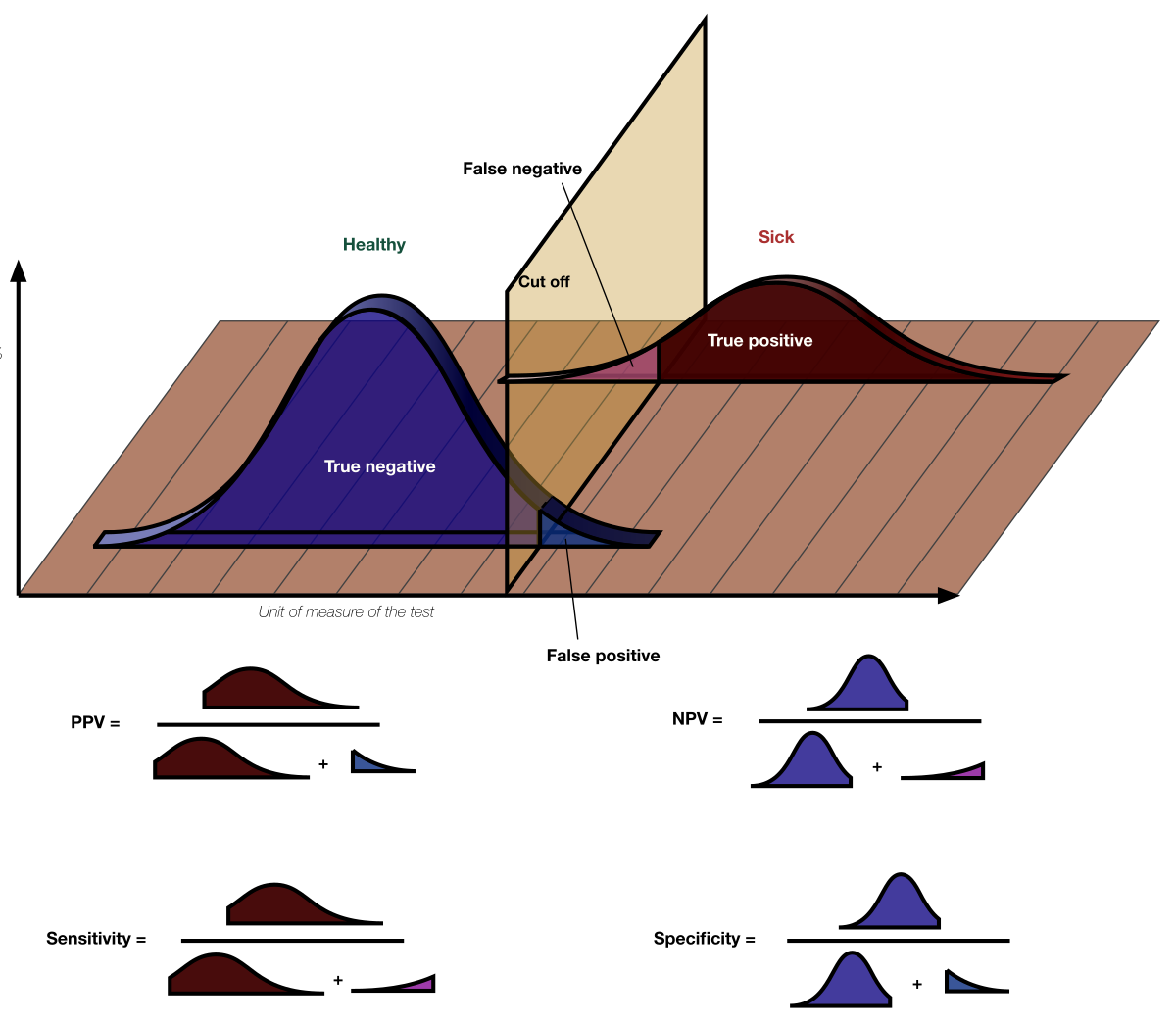
I want to do an evaluation of a pattern search tool. For that I want to calculate sensitivity and specificity.
How can I determine the true positives and false positives, the real pattern (verified experimentally) are not all known. And how can I calculate the sensitivity and specificity.
Cordially
I want to do an evaluation of a pattern search tool. For that I want to calculate sensitivity and specificity.
How can I determine the true positives and false positives, the real pattern (verified experimentally) are not all known. And how can I calculate the sensitivity and specificity.
Cordially

Comment