Hello,
I would love some advice on my linked image below. I was using FastQC to look at the data from a recent MiSeq run. All my isolates are flagged at the "Per base sequence content". I am to understand that a 'good' graph is expected to look like all the bases are of similar % throughout. The DNA sequenced is from Pseudomonas aeruginosa, and I am seeing a high GC content (as expected) and much lower AT content.
Any advice or explanation would be greatly helpful!
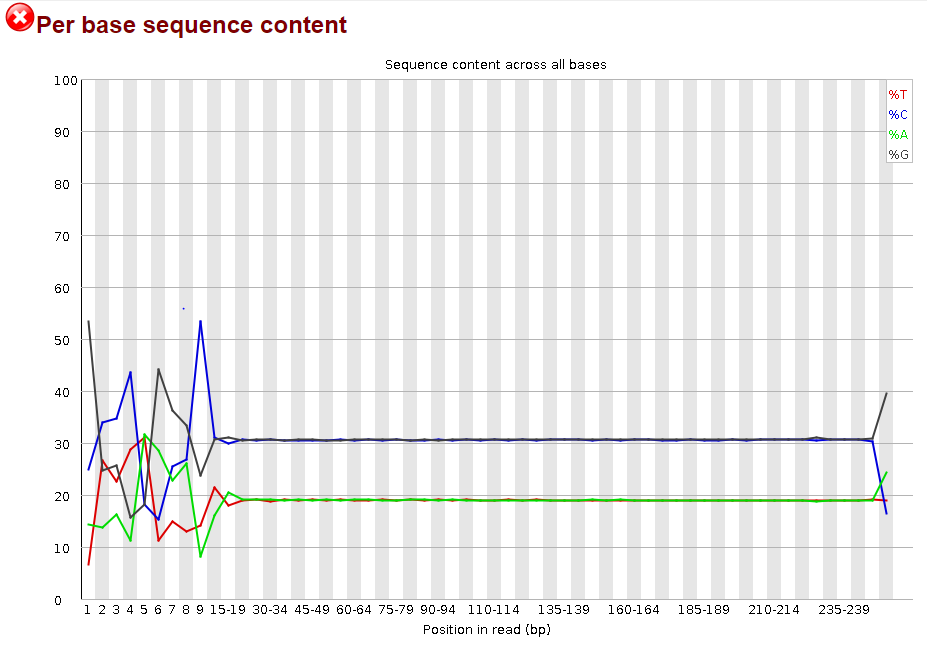
I would love some advice on my linked image below. I was using FastQC to look at the data from a recent MiSeq run. All my isolates are flagged at the "Per base sequence content". I am to understand that a 'good' graph is expected to look like all the bases are of similar % throughout. The DNA sequenced is from Pseudomonas aeruginosa, and I am seeing a high GC content (as expected) and much lower AT content.
Any advice or explanation would be greatly helpful!
Comment