I gave the Illumina TruSeq RNA Prep Kit v2 an initial go with two samples. I followed the Illumina protocol exactly, starting with 1 μg of HeLa cell RNA purified from Qiagen RNeasy columns.
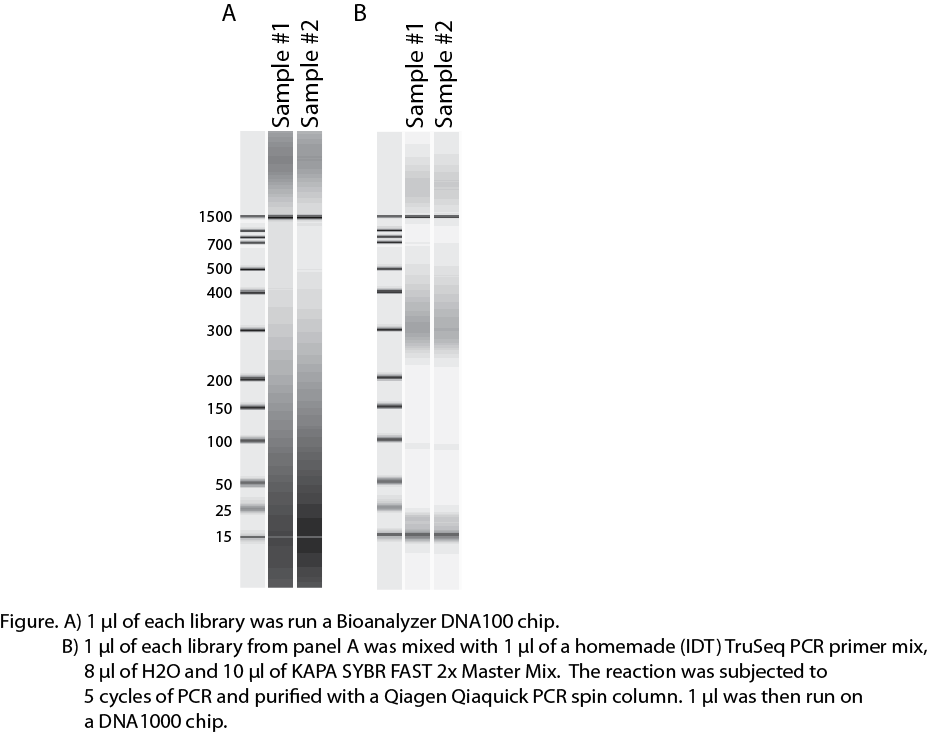
I ran 1 ul of the library after amplification on a Bioanalyzer DNA1000 chip got a really strange image (see attached image A). I really can't make sense of what is going on here. My only thought was that it was some weird result of over-amplification so I took 1 ul of the library, mixed it with 1 ul of a homemade (IDT) PCR Primer Cocktail, 8 ul of H2O and 10 ul of Kapa SYBR FAST 2X master mix. I then subjected the sample to 5 cycles of PCR, purified the reaction on a Qiagen Qiaquick PCR purification spin column and ran 1 ul of that on another DNA1000 chip (see attached image B).
In the re-amplification the library looks better. There are some higher molecular weight fragments even though you can still see that there are plenty of primers left in the reaction. But I would still guess that the larger material is DNA fragments annealing by their adapter sequences.
Anyway, my question is has anyone ever seen a Bioanalyzer image like this? Did you sequence the library? What is your guess to what is going on?
I ran 1 ul of the library after amplification on a Bioanalyzer DNA1000 chip got a really strange image (see attached image A). I really can't make sense of what is going on here. My only thought was that it was some weird result of over-amplification so I took 1 ul of the library, mixed it with 1 ul of a homemade (IDT) PCR Primer Cocktail, 8 ul of H2O and 10 ul of Kapa SYBR FAST 2X master mix. I then subjected the sample to 5 cycles of PCR, purified the reaction on a Qiagen Qiaquick PCR purification spin column and ran 1 ul of that on another DNA1000 chip (see attached image B).
In the re-amplification the library looks better. There are some higher molecular weight fragments even though you can still see that there are plenty of primers left in the reaction. But I would still guess that the larger material is DNA fragments annealing by their adapter sequences.
Anyway, my question is has anyone ever seen a Bioanalyzer image like this? Did you sequence the library? What is your guess to what is going on?

Comment