hello, I would like to know if there is a way to determine whether two set of data from rna seq are significantly different, like these:
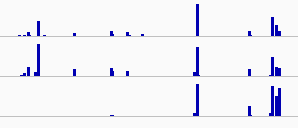
for example, the first two rows are similar, while the third is only half similar. The two data sets are represented as arrays [0,0,1,9,4....]
any suggestion? thanks
for example, the first two rows are similar, while the third is only half similar. The two data sets are represented as arrays [0,0,1,9,4....]
any suggestion? thanks