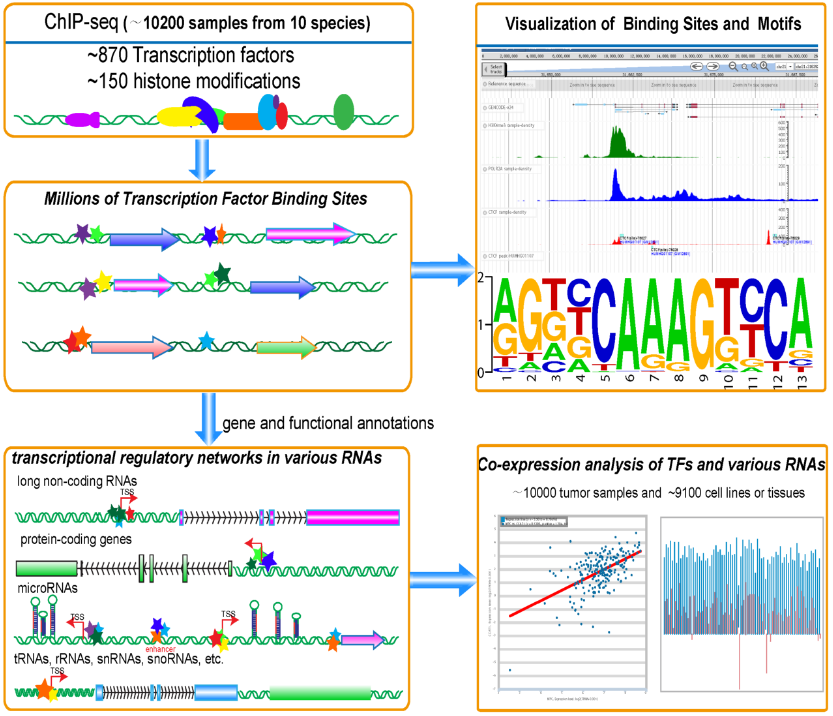
ChIPBase v2.0 (http://rna.sysu.edu.cn/chipbase/) has been developed to explore the regulatory networks between transcription factors and genes (proteins, lncRNAs, miRNAs, snoRNAs, tRNAs... ). ChIPBase v2.0 integrated ~10,200 curated ChIP-seq datasets. We identified thousands of Binding Motif Matrices and their binding sites from ChIP-seq data of DNA-binding proteins and predicted millions of transcriptional regulatory relationships between transcription factors and genes. We constructed “Regulator” module to predict hundreds of transcription factors that were involved in or affected transcription of ncRNAs and protein-coding genes. Moreover, we built a web-based tool, Co-Expression, to explore the co-expression patterns between DNA-binding proteins and various types of genes by integrating the gene expression profiles of ~10,000 tumor samples and ~9,100 normal tissues or cell lines. ChIPBase also provides a ChIP-Function tool and a genome browser to predict functions of diverse genes and visualize various ChIP-seq data. This study will greatly expand our understanding of the transcriptional regulations of ncRNAs and protein-coding genes.