RMBase (RNA Modification Base) is a RNA modification database and designed for decoding the landscape of RNA modifications (>100 different RNA modification types) identified from 404 high-throughput RNA sequencing data (Pseudo-seq, Ψ-seq, CeU-seq, Aza-IP, MeRIP-seq, m6A-seq, miCLIP, m6A-CLIP, RiboMeth-seq).
It contains ~124200 N6-Methyladenosines (m6A), ~9500 pseudouridine (Ψ) modifications, ~1000 5-methylcytosine (m5C) modifications, ~1210 2′-O-methylations (2′-O-Me) and ~3130 other types of RNA modifications.
RMBase demonstrated thousands of RNA modifications located within regulatory RNAs (e.g. mRNAs, lncRNAs, miRNAs, pseudogenes, circRNAs, snoRNAs etc.), miRNA target sites and disease-related SNPs.
This platform will greatly help biologists to explore the prevalence, mechanism and function of various RNA modifications and link them with human cancers and other diseases.
RMBase is freely available at http://rna.sysu.edu.cn/rmbase/.
RMBase paper is available at http://nar.oxfordjournals.org/content/44/D1/D259
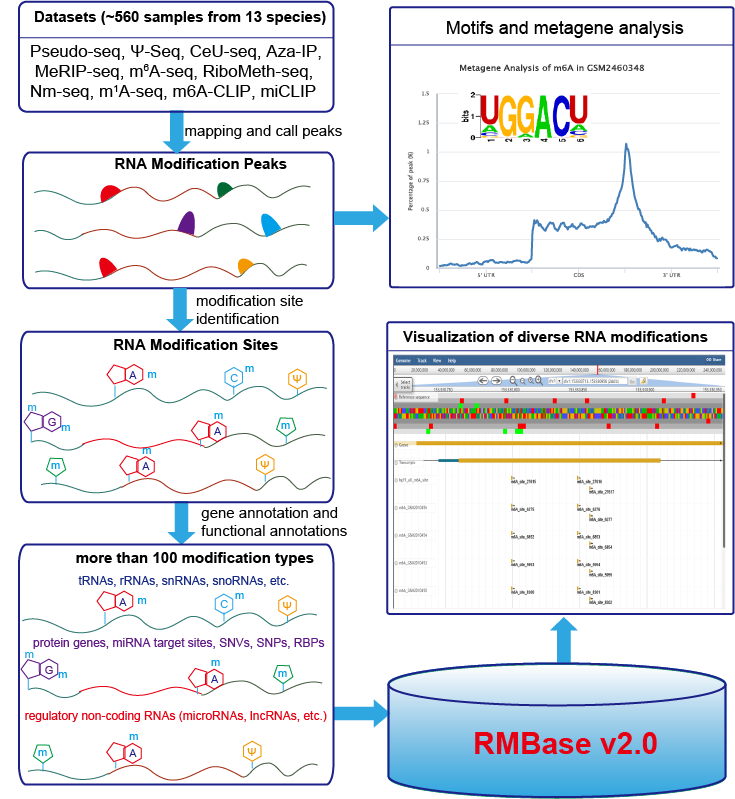
It contains ~124200 N6-Methyladenosines (m6A), ~9500 pseudouridine (Ψ) modifications, ~1000 5-methylcytosine (m5C) modifications, ~1210 2′-O-methylations (2′-O-Me) and ~3130 other types of RNA modifications.
RMBase demonstrated thousands of RNA modifications located within regulatory RNAs (e.g. mRNAs, lncRNAs, miRNAs, pseudogenes, circRNAs, snoRNAs etc.), miRNA target sites and disease-related SNPs.
This platform will greatly help biologists to explore the prevalence, mechanism and function of various RNA modifications and link them with human cancers and other diseases.
RMBase is freely available at http://rna.sysu.edu.cn/rmbase/.
RMBase paper is available at http://nar.oxfordjournals.org/content/44/D1/D259

Comment