Originally posted by dnusol
View Post
Seqanswers Leaderboard Ad
Collapse
Announcement
Collapse
No announcement yet.
X
-
Maybe RNAseq is biased by nature?
Check these other threads
Application of sequencing to RNA analysis (RNA-Seq, whole transcriptome, SAGE, expression analysis, novel organism mining, splice variants)
Discussion of any scientific study related to high content or next generation genomics. Whole genome association, metagenomics, digital gene expression, etc.
Leave a comment:
-
Hi, rhinoceros, here are more details about the samples.Originally posted by rhinoceros View PostLooks to me like the reads come from two very different sources (the two peaks). Perhaps even three sources.. I don't know. How are we supposed to tell since you didn't specify what was sequenced. Since the read counts (of the peaks) are somewhat similar, perhaps your source has two chromosomes, which have different GC (not unheard of).
The total RNA was extracted from one piece of mouse muscle by Qiagen RNeasy fibrous tissue mini kit. The quality of RNA were estimated by Agilent bioanalyzer. A260/A280 for all samples were around 2.0. RIN for all RNAs were around 10. It seems the quality of RNAs is excellent. And I didn't seem a chance to have other chromosomes.
The Libraries were prepared by a RNAseq center with an illumina TruSeq kit, but I know little about that...
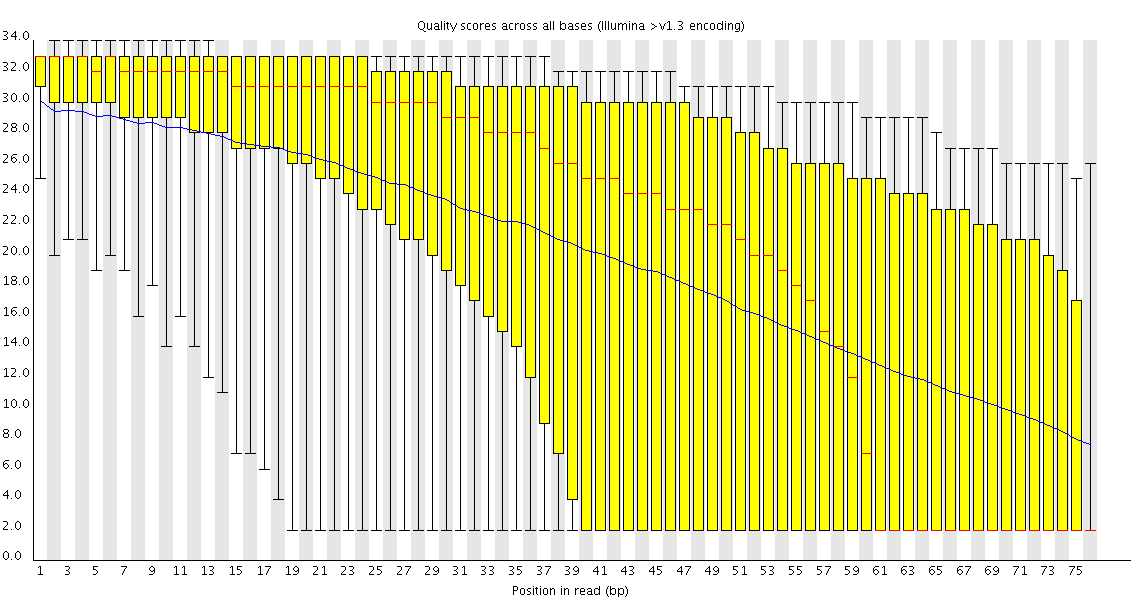
Here is the whole FastQC report, which may give you more information. http://zgan.weebly.com/uploads/8/9/3...tqc_report.zip
More clues for me? ThanksLast edited by ZoeG; 06-18-2013, 03:53 PM.
Leave a comment:
-
Thanks, dnusol. I went through the webpage, what I can tell is that the library seems something wrong. The quality score actually seems okay. When I mapped the data to its reference, >90% reads were mapped successfully. Do this result conflict with the conclusion of wrong library? Confused..Originally posted by dnusol View Post
Leave a comment:
-
Looks to me like the reads come from two very different sources (the two peaks). Perhaps even three sources.. I don't know. How are we supposed to tell since you didn't specify what was sequenced. Since the read counts (of the peaks) are somewhat similar, perhaps your source has two chromosomes, which have different GC (not unheard of).Last edited by rhinoceros; 06-14-2013, 04:23 AM.
Leave a comment:
-
This link might be useful
Leave a comment:
-
Contamination? What does this per sequence GC content mean?
Hello, I am quite new to RNAseq analysis.
When I did the quality control for my Illumina HiSeq data, I found an abnormal per sequence GC content, as the picture shows.
It seems some contamination?
Need help, thanksTags: None
Latest Articles
Collapse
-
by seqadmin
Spatial biology is an exciting field that encompasses a wide range of techniques and technologies aimed at mapping the organization and interactions of various biomolecules in their native environments. As this area of research progresses, new tools and methodologies are being introduced, accompanied by efforts to establish benchmarking standards and drive technological innovation.
3D Genomics
While spatial biology often involves studying proteins and RNAs in their...-
Channel: Articles
Yesterday, 07:30 PM -
-
by seqadmin
Many organizations study rare diseases, but few have a mission as impactful as Rady Children’s Institute for Genomic Medicine (RCIGM). “We are all about changing outcomes for children,” explained Dr. Stephen Kingsmore, President and CEO of the group. The institute’s initial goal was to provide rapid diagnoses for critically ill children and shorten their diagnostic odyssey, a term used to describe the long and arduous process it takes patients to obtain an accurate...-
Channel: Articles
12-16-2024, 07:57 AM -
ad_right_rmr
Collapse
News
Collapse
| Topics | Statistics | Last Post | ||
|---|---|---|---|---|
|
Started by seqadmin, 12-30-2024, 01:35 PM
|
0 responses
21 views
0 likes
|
Last Post
by seqadmin
12-30-2024, 01:35 PM
|
||
|
Started by seqadmin, 12-17-2024, 10:28 AM
|
0 responses
41 views
0 likes
|
Last Post
by seqadmin
12-17-2024, 10:28 AM
|
||
|
Started by seqadmin, 12-13-2024, 08:24 AM
|
0 responses
55 views
0 likes
|
Last Post
by seqadmin
12-13-2024, 08:24 AM
|
||
|
Started by seqadmin, 12-12-2024, 07:41 AM
|
0 responses
40 views
0 likes
|
Last Post
by seqadmin
12-12-2024, 07:41 AM
|

Leave a comment: